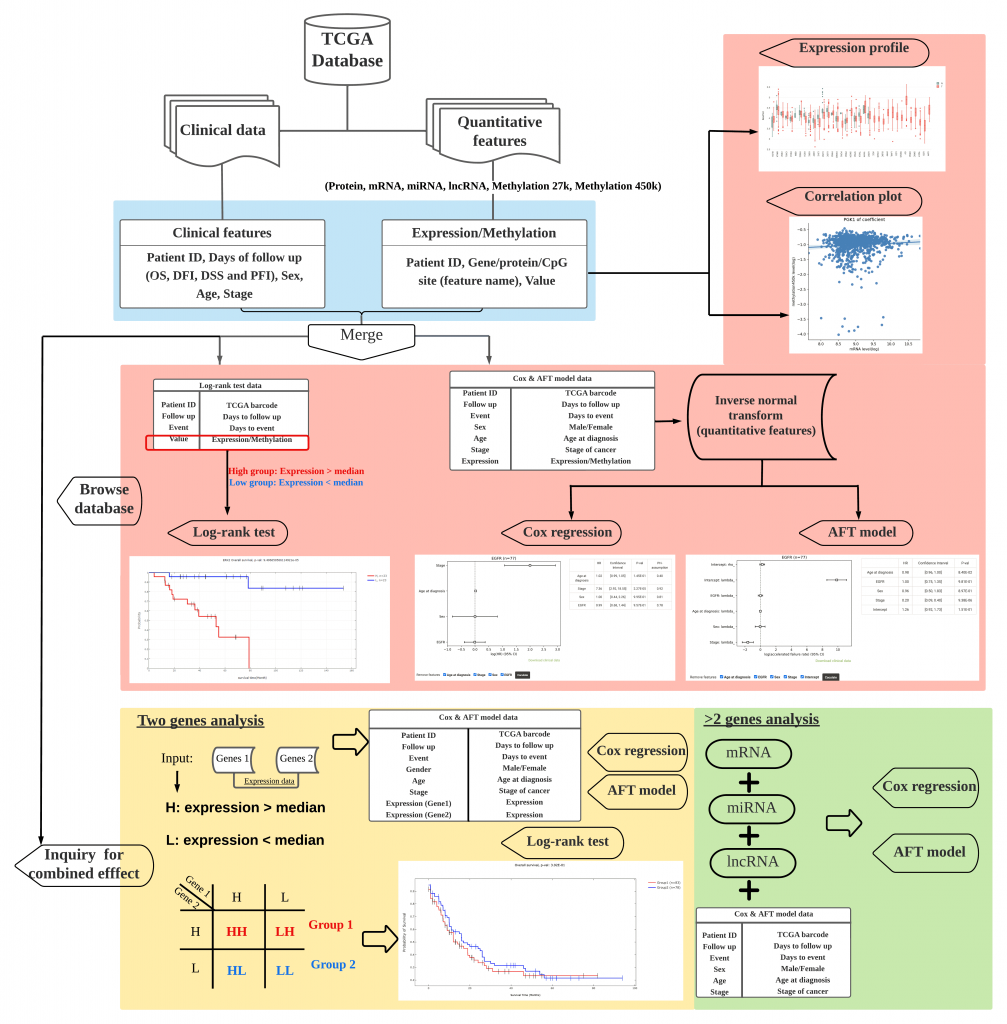
DoSurvive
DoSurvive provides univariate correlation analysis results including mRNA, miRNA, lncRNA, protein and Methylation and cancer patient survival. Users can also use TCGA data online to perform multivariate survival analysis in real time.
In addition, DoSurvive provides three online survival analysis services including Log-rank test, Cox regression and AFT, allowing users to analyze their own data (when the data violates Proportional Hazard, Log-rank test or AFT can be used for multi-variable analysis).
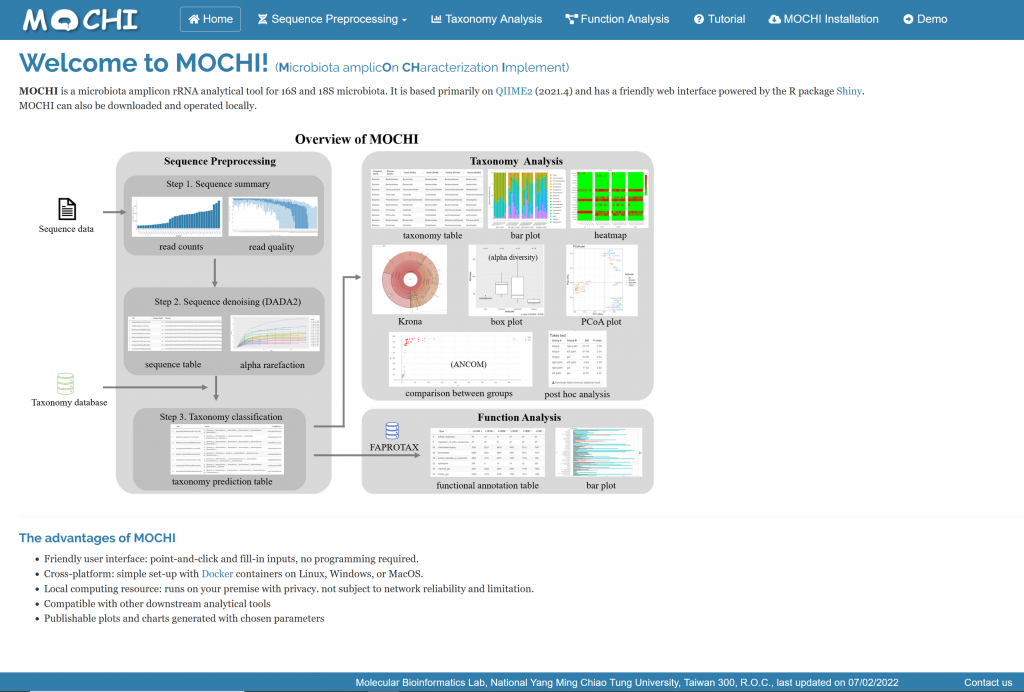
MOCHI
MOCHI is a microbiota amplicon rRNA analytical tool for 16S and 18S microbiota. It is based primarily on QIIME2 (2021.4) and has a friendly web interface powered by the R package Shiny.
1. Friendly user interface: point-and-click and fill-in inputs, no programming required.
2. Cross-platform: simple set-up with Docker containers on Linux, Windows, or MacOS.
3. Local computing resource: runs on your premise with privacy. not subject to network reliability and limitation.
4. Compatible with other downstream analytical tools
5. Publishable plots and charts generated with chosen parameters
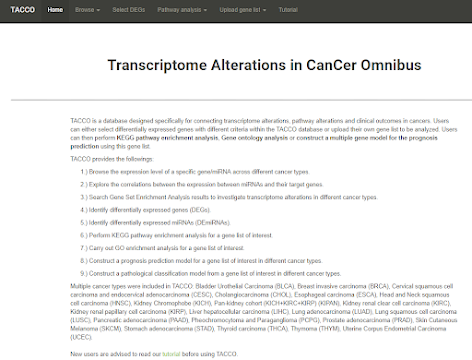
TACCO
This database provides information regarding the 25 cancers in TCGA, including:
1. Differential expression of RNA and microRNA
2. Analysis of the reaction path involved in the above differential expression RNA
3. A module for predicting cancer cluster
4. The prediction module that can predict the prognosis of cancer
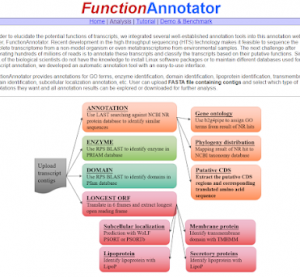
FunctionAnnotator
This webpage tool combines multiple prediction tools to provide functional annotations on the contigs formed after transcript sequencing of non-model species. The prediction analysis items include membrane proteins, enzyme proteins, subcellular localization, GO (Gene ontology) function annotations, etc.
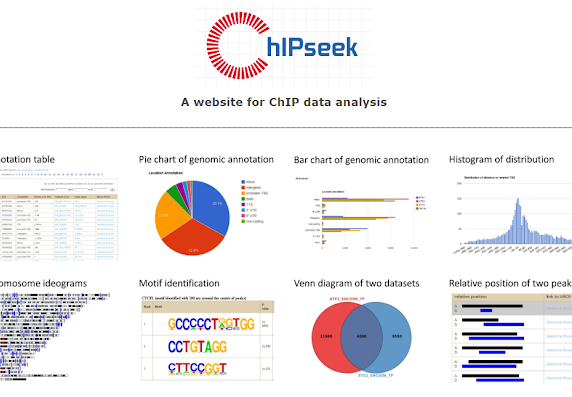
CHIPSeek
The bioinformatics webpage tool is for assisting in the analysis of ChIP (Chromatin Immunoprecipitation) data, providing peak annotation, binding motif prediction, peak region comparison, peak filtering and other functions.
CHLOROMITOCU
This is a database specially constructed for the analysis of the codon usage frequency of chloroplast and mitochondria. In addition to providing data query, users can also upload sequences for analysis by themselves, and provide a comparison of codon usage frequency.
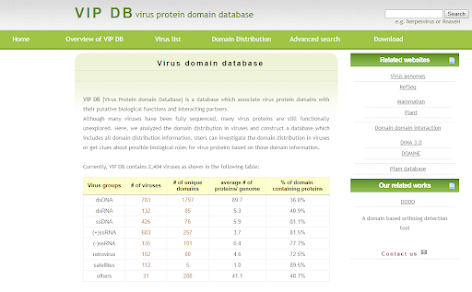
VIP DB
This is a database that specifically analyzes and compares the distribution of protein domains in viruses. Users can find out the use of a specific domain in viruses, plants, animals, or of two domains that have or have not ever appeared together in the same viral gene.